Relationship extraction from unstructured species descriptions using TaxonNERD and LLaMA2 - 7B
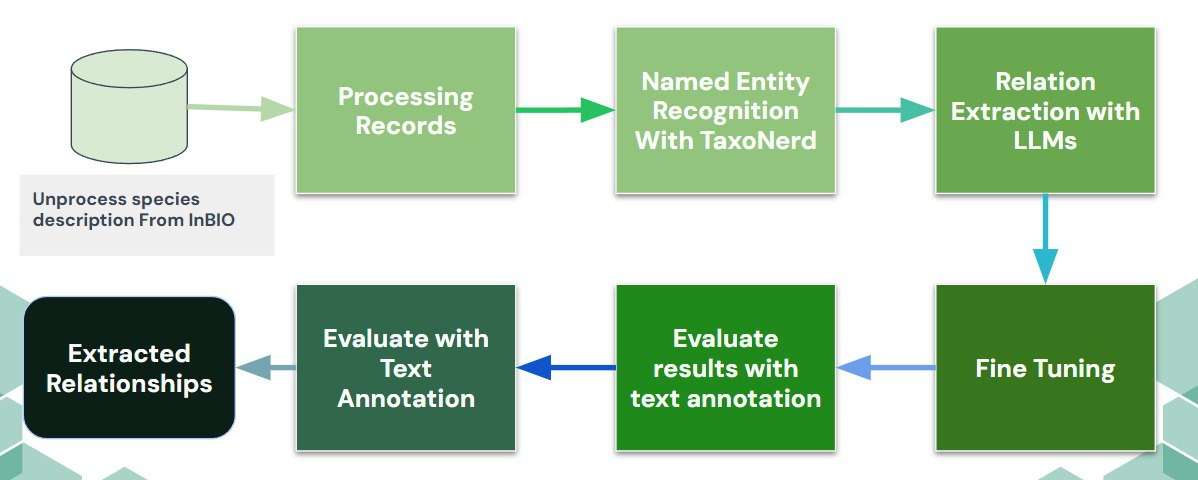
This project explores how to extract structured information on feeding relationships between animal species from originally unorganized textual descriptions. The project combines ecological ontologies with advanced Natural Language Processing (NLP) methods, using specialized tools such as TaxoNERD for entity recognition, and the LLaMA2-7b model for extracting specific relationships, particularly predator-prey dynamics. The dataset used comes from the National Institute of Biodiversity (INBio), with descriptions in Spanish and English about feeding habits and ecological behaviors.
The methodology consisted of preprocessing INBio texts to identify taxonomic entities and feeding-related terms using TaxoNERD. Subsequently, the LLaMA2-7b model was employed to extract concrete relationships, thus generating a structured dataset where the diet of each studied species is clearly detailed. The obtained results were manually reviewed through a custom text annotation tool developed specifically for this project, ensuring the quality and accuracy of the generated information.
The evaluation showed promising results, with a precision of 68%, recall of 73%, and F1-score of 71%, particularly highlighting the model's ability to detect key relationships in species descriptions. As a next step, the model will be further refined to improve its accuracy and broaden its scope to other ecological relationships, thereby strengthening its usefulness for future biodiversity research and environmental conservation efforts.
Optimization of language models for species information retrieval and generation: Integrating contextual knowledge through the Plinian Core standard
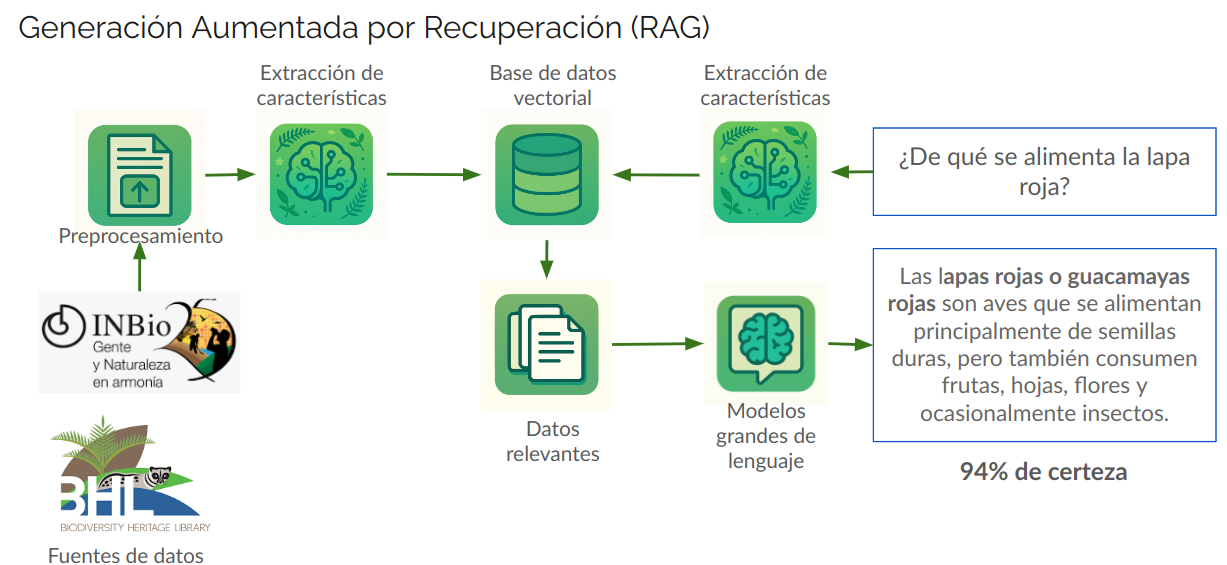
This project aims to leverage the power of artificial intelligence to answer biodiversity-related questions in Spanish clearly and reliably. Through an approach called Retrieval-Augmented Generation (RAG), data from sources such as the National Institute of Biodiversity of Costa Rica (INBio) and the Biodiversity Heritage Library (BHL) are integrated with optimized language models. This allows the system, when asked a question such as “What does the scarlet macaw eat?”, to retrieve precise information from biological descriptions enriched with context in Plinian Core format (from the vector database), process it, and generate a response using large language models, accompanied by a certainty level. In this way, an innovative tool is built to bring scientific knowledge about species in a region closer to society, facilitating access to validated and useful information for education, research, and biodiversity conservation. More information about the project is available at https://aclanthology.org/2024.naacl-industry.31/ and https://github.com/biodiversidad-itcr.
Monitoring species in Costa Rica through camera traps and transformers
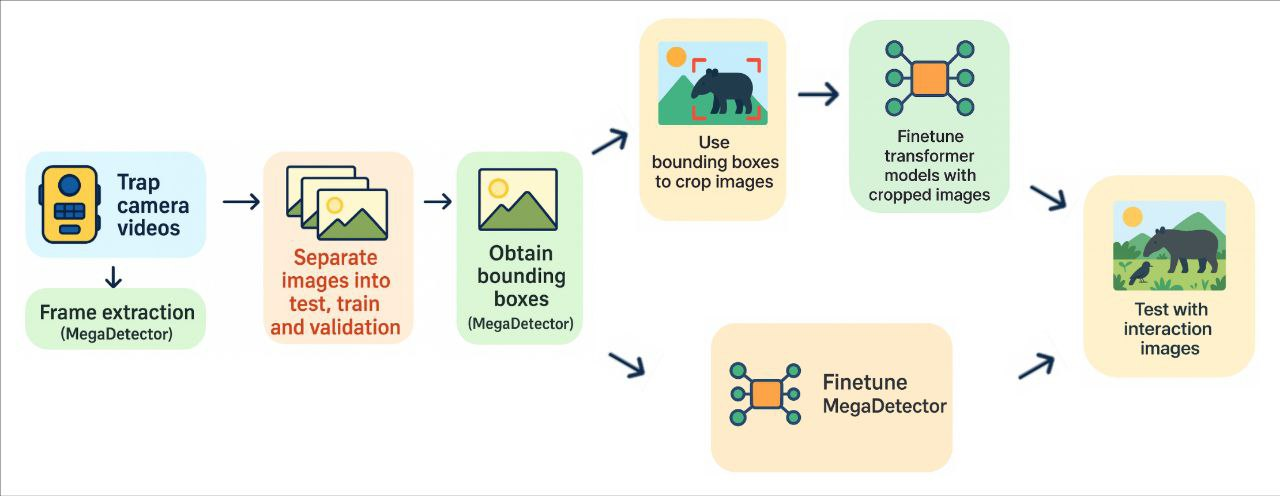
The main goal of this project is to facilitate the identification of animal species in Costa Rica, specifically those approaching the waterhole at CEMEDE in the National University of Costa Rica, using videos captured by camera traps. To this end, experiments were conducted with image classification models based on transformer architectures (DeiT, Swin-S, and Efficient-ViT), chosen for their high potential accuracy and limited application in this type of context. Additionally, the MegaDetector model, recognized for its animal detection capabilities in images, was adapted to optimize its performance on the specific classes of this study.
The dataset was built from approximately 26,000 images extracted from the videos, corresponding to 44 animal classes. Due to class imbalance, an undersampling process had to be applied to obtain between 100 and 500 images of 12 classes, excluding endangered species to ensure their protection. The final dataset, composed of about 6,000 images, was processed with the basic MegaDetector to extract and crop the regions where animals appeared in each image. These cropped images were then used to train the three selected transformer models and the adjusted MegaDetector, after which their performance in the classification task was compared.
Ecosystem Simulator Using Large Language Models (LLM).
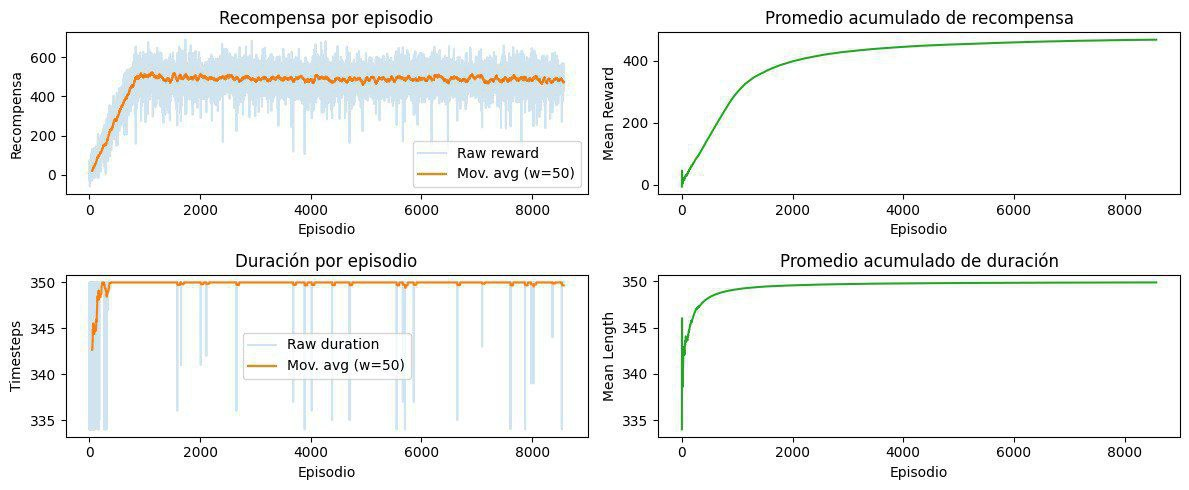
This project seeks to develop an ecosystem simulator that models the interaction between multiple agents, resources, and the environment, using Deep Reinforcement Learning combined with advanced natural language models (LLM). Initially, a simplified 2D environment will be implemented using Python's Gym library, where various agents will learn optimal survival strategies through algorithms such as DQN, PPO, and A2C. In addition, advanced reasoning and interaction capabilities will be incorporated through an LLM (such as LLaMA 3), enabling intelligent decision-making, natural communication between agents, and generating understandable explanations of their actions.
Subsequently, the possibility of scaling the simulator to a three-dimensional environment using Minecraft together with the MineRL library will be evaluated, exploring technical feasibility and documenting possible limitations or challenges. During development, several deliverables will be produced, including a complete code repository, technical reports on the architecture and algorithms used, as well as visual demonstrations of agent performance in both environments (2D and 3D).
Finally, this project aims to offer not only a versatile simulation environment rich in biological interactions but also a robust platform to investigate emergent behaviors in multi-agent systems and explore innovative applications of artificial intelligence and natural language processing. The simulator will allow deeper studies of ecological dynamics while facilitating future research on the practical and effective integration between deep reinforcement learning and advanced generative language models.
